‘Spare tire genes’ explain why some genes can be lost by cancer cells
Blog
- Student Profile: Mahmoud Said El-Sayed Abd-Allah
- Can election results be predicted by correcting biases in social polls from X?
- International Women’s Day: Celebrating Ad Astra Scholar Ava Canning
- Chinese New Year 2024 Celebration
- MSc Advanced Software Engineering Alumni
- Generative AI in Computing Education: Wrecking Ball or Holy Grail?
- My internship in digital strategy at Limerick City & County Council
- Clown Computing 101
- International Men's Day 2023
- Top marks for ChatGPT in the Leaving Certificate Computer Science Examination
- Alumnus Interview
- Student profile: Pasika Ranaweera, PhD student
- Staff profile: Associate Professor Neil Hurley, Head of School
- UCD CS PhD candidate award from IEEE Consumer Electronics Magazine
- UCD-Insight Collaboration Wins Prestigious Publication Award
- W@CS Alumni Roundtable
- Staff profile: Dr. Fatemeh Golpayegani
- Interning at a Smaller Tech Company
- Powering through the pandemic: My Remote Research Internship Experience
- Exploring Sense of Belonging in Computer Science Students
- Student Inter-Society Tech & Enterprise Meetup (SISTEM) held in UCD
- Computer Science Research and the COVID -19 Pandemic
- Zoom fatigue: how to make video calls less tiring
- SIGCSEire Launched at UCD CS
- Best Paper at the International Conference on Case-Based Reasoning (2019)
- ‘Spare tire genes’ explain why some genes can be lost by cancer cells
- Bi-annual CS Graduate Research Symposium
- UCD CS Postdoctoral fellow Claudia Mazo selected as a member of the ACM Future of Computing Academy
- Security, Privacy and Digital Forensics in the Cloud
- Chidubem Iddianozie: PhD student and GitHub Ambassador in UCD
- UCD CS PhD student selected to attend the Heidelberg Laureate Forum
- New Project: Evidence-Based Decision Support for Real-Estate Investment
- UCD projects celebrate Europe Day
- Research Award at the 2019 ACM SIGCSE Technical Symposium on Computer Science Education
- Top Tips for Student Scholarships
- I am a Computer Scientist and a Cancer Biologist
- Critical thinking and data ethics in UCD CS
- Teaching at BDIC Beijing
- Reading 35,000 Books
- Secret to a Great Internship
- 12 Tips for PhD Researchers
- Buddy Coders - a new initiative to support women in Computer Science

The majority of our approximately 20,000 genes can be mutated or deleted in cancer cells without killing these cells. This is surprising given the key role that genes play in our cells - genes encode the instructions to build molecules called proteins, which form the machinery that carries out many of the functions of our cells. In a recent study we asked: which genes can be lost in cancer cells, and why?
We addressed these questions by analysing data on which genes are required for the survival of over 500 different cancer (opens in a new window)cell lines. Cancer cell lines are cells taken from patient tumours and grown in the laboratory. Recent technical advances have made it possible to individually delete each gene in the human genome in many different cancer cell lines. One such advance is the development of (opens in a new window)CRISPR gene editing methods, which can be used to cut DNA with a molecular scissors to remove a particular gene. This allows us to determine whether a given gene is essential for the survival of the cells in a given cancer cell line.
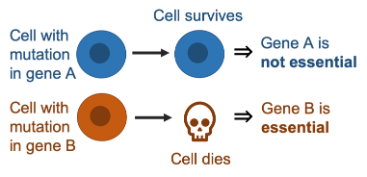
Whether or not a gene is essential has often been treated as a static, binary property, but it has become apparent that gene essentiality can vary significantly across different genetic backgrounds. This means that a particular gene can be essential for one cancer cell line but not another.
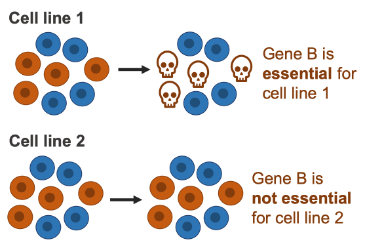
From the (opens in a new window)DepMap project we obtained the results of screening, with the use of CRISPR, 558 different cancer cell lines, which originate from more than 20 different cancer types. We used these to call each of ~16,000 genes either essential or not essential in each cell line, and then calculated the percentage of cell lines in which each gene was essential.
We found that paralog genes are, in general, essential in fewer cancer cell lines than genes that are not paralogs (these are called singletons). Paralogs are genes that have been duplicated at some point in evolutionary history, resulting in our genome having two copies of (roughly) the same gene—a paralog pair. These paralog pairs, which make up over half the genes in our genome, are a potential source of redundancy, similar to a car having a spare tire. Accordingly, one explanation for why paralogs are less often essential is that their loss, which could be likened to a flat tire for the cell, can be mitigated by a ‘spare tire gene’. We’ll refer to the two genes that form a paralog pair as A1 and A2, where A2 could be considered the ‘spare tire gene’ for A1.
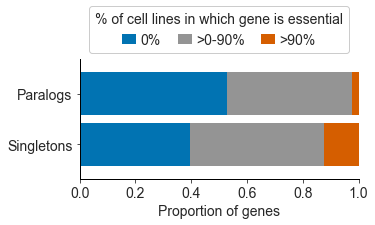
We expected that while the loss of one gene from a paralog pair could be tolerated, due to the existence of a ‘spare tire gene’, the loss of both members would not be tolerated - i.e. the car would cease to function if it lost both a tire (A1) and its spare (A2).

To test this we investigated 1,819 paralog pairs where the A1 gene was essential in some but not all of the cancer cell lines (i.e. it falls into the grey category of the bar graph above). This allowed us to compare the status of the A2 gene in cell lines where A1 is and is not essential. Specifically, we compared the expression of A2, which provides a relative measure of the amount of A2 that is present in the different cell lines. For 238 (~13%) of the paralog pairs, we found that the expression of A2 was on average significantly lower in the cell lines where A1 is essential compared to the cell lines where A1 is not essential. Thus the amount of A2 that is present can explain why loss of A1 is tolerated by some cell lines and not others. This is indicative of a phenomenon called synthetic lethality - the loss of A1 alone can be tolerated, but the simultaneous loss of A1 and A2 is not tolerated.
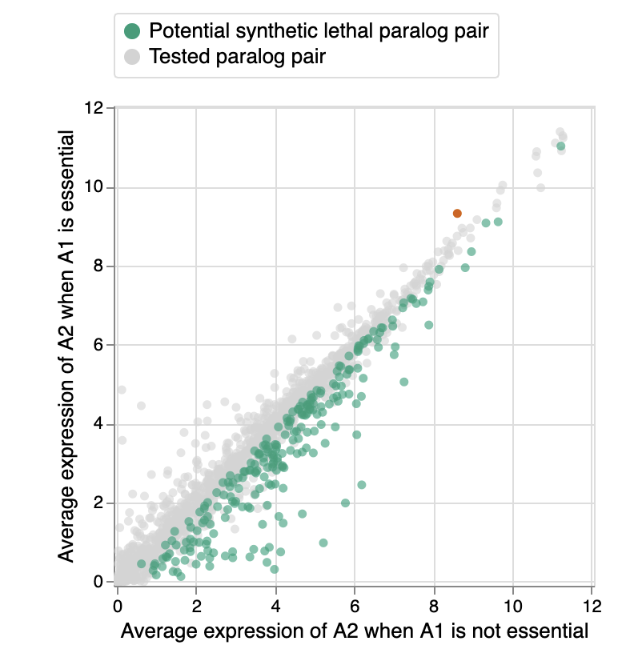
Synthetic lethality represents a promising approach for targeted therapies in cancer, in part because gene loss is common in cancer cells, but rare in healthy cells. If we know a cancer cell has lost one member of a synthetic lethal pair, e.g. a ‘spare tire gene’, we can anticipate that it will be particularly vulnerable to loss of the other member. In this study we show that ~13% of paralogs exhibit such a synthetic lethal relationship, and that this explains why loss of some genes is sometimes, but not always, tolerated by cancer cells.
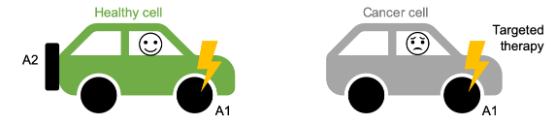
Although the focus of our work was not the identification of new targets for cancer therapies, our findings suggest that certain types of paralogs are more likely to be involved in synthetic lethal relationships than others. This may ultimately help guide the search for new therapeutic targets.
Research team:
This study was carried out by Barbara De Kegel, a PhD student in Systems Biology Ireland / the UCD School of Computer Science, and Colm J. Ryan, an assistant professor in the UCD School of Computer Science.
You can read the full paper here: (opens in a new window)https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1008466